Section: New Results
Sparse 3D reconstruction in fluorescence imaging
Participants : Emmanuel Soubies, Laure Blanc-Féraud, Sébastien Schaub, Gilles Aubert.
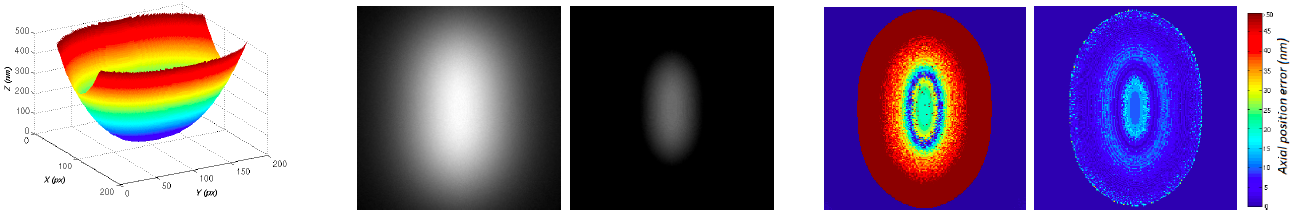
Sparse reconstruction Super-resolution microscopy techniques allow to overstep the diffraction limit of conventional optics. Theses techniques are very promising since they give access to the visualisation of finer structures which is of fundamental importance in biology. In this work we deal with Multiple-Angle Total Internal Reflection Microscopy (MA-TIRFM) which allows reconstructing 3D sub-cellular structures of a single layer of behind the glass coverslip with a high axial resolution. The 3D volume reconstruction from a set of 2D measurements is an ill-posed inverse problem and requires some regularization. Our aim in this work is to propose a new reconstruction method for sparse structures that is robust to Poisson noise and background fluorescence. The sparse property of the solution can be seen as a regularizer using the -norm. Let us denote the unknown fluorophore density, then the problem states as
where is defined from the likelihood function of the observation given , is a weight parameter and denotes the -norm (which counts the number of nonzero components of ). In order to solve this combinatorial problem, we propose a new algorithm based on a smoothed -norm allowing minimizing the non-convex energy (1 ). Following [20] , the idea is to approach the -norm by a suitable continuous function depending on a positive parameter and tending to the -norm when the parameter tends to zero. Then the algorithm solves a sequence of functionals which starts with a convex one (on a large convex set) and introduce progressively the non-convexity of the -norm (Graduated Non Convexity approach). Figure 1 shows the accuracy of the method on a simulated membrane.
|
Axial profile calibration In order to turn on real sample reconstructions we need to perform a calibration of the TIRF microscope. Its principle is based on an evanescent wave with an exponential theoretical decay. However this decay is generally not a pure exponential in practice and we need to have a good knowledge about it. Then based on a phantom specimen of known geometry (bead) we are working on a method to estimate experimentally/numerically this decay profile and calibrate all parameters of the system.